Document search using Claude and an inverted index.
Soooo vector databases are pretty popular right now for document search, but it has some drawbacks: choosing the right embedding models, finding the right chunk size, indexing into high dimensional embeddings and managing specialized infrastructure.
This complexity might be unnecessary for some use cases that just have a bunch of docs in S3 and want to RAG over them.
I started with the thesis that LLM generated query sets combined with a good index can match vectorDB performance.
The idea
I decided to implement the simplest idea:
- I used an LLM (Claude Sonnet 4.5 in this case) to generate possible keywords that could be present in documents related to the search query. I played around with a few numbers until I landed on 10 query sets each containing 2 keywords. Looking at an example query “what has been published about COVID-19 mRNA vaccines”:
Set 1: ['mrna', 'vaccine']
Set 2: ['coronavirus', 'immunization']
Set 3: ['covid', 'vaccination']
Set 4: ['messenger', 'inoculation']
Set 5: ['spike', 'protein']
Set 6: ['pandemic', 'prophylaxis']
Set 7: ['pfizer', 'moderna']
Set 8: ['antibody', 'response']
Set 9: ['viral', 'prevention']
Set 10: ['immunity', 'development']
The LLM generates both obvious combinations (‘mrna’ + ‘vaccine’) and creative variations (‘messenger’ + ‘inoculation’, ‘pfizer’ + ‘moderna’). This diversity is key to matching vector search performance.
- These searches were then executed against an inverted index. Again I experimented a bit and landed on ‘AND’ within each set, ‘OR’ across sets. This helps control retreival size while allowing creativity.
- The retrieved documents were then ranked using BM25.
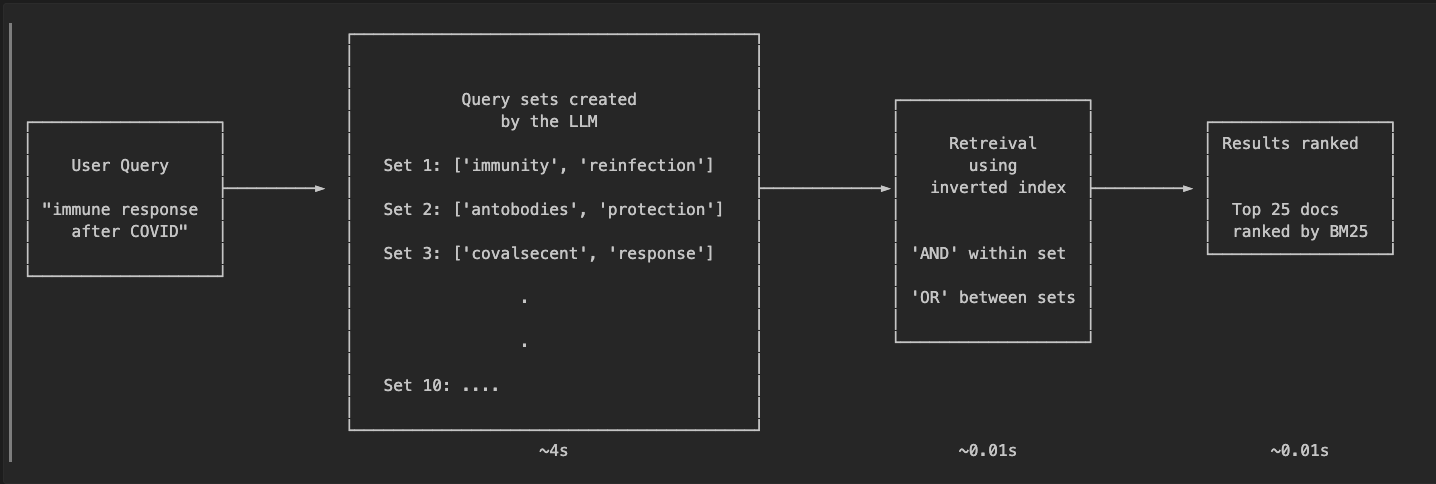
This gives us the semantic understanding of an LLM combined with the speed of traditional search. The LLM has a reasonable understanding of the user intent and can generate diverse query sets.
Dataset Selection
I chose a dataset with lots of documents, relatively short content. The TREC-COVID dataset from the BEIR benchmark was ideal:
- ~171k documents.
- Typically under 1,000 words per document because they contain abstracts.
- Somewhat domain specific as it contains biomedical literature articles. (This is good for Information Retrival tests).
Note: I’m curious if there are other datasets that might be better suited to this IR task. Happy to run this against more datasets.
Procedure
The BEIR benchmark has a mapping of which documents are relevant for each query. When the query is sent to different Information Retrieval (IR) systems the retrieved results are compared with the known mappings to compute different metrics.
These are the metrics we will be looking at:
- NDCG (Normalized Discounted Cumulative Gain): Measures ranking quality by rewarding relevant documents that appear higher in results, with scores from 0-1 where 1 is perfect ranking.
- Recall@k: The fraction of all truly relevant documents that appear in the top-k results (e.g., Recall@100 of 0.01 means 1% of relevant docs were found in top 100).
- MAP (Mean Average Precision): Averages the precision scores at each position where a relevant document is found, rewarding systems that rank relevant docs consistently high.
- MRR (Mean Reciprocal Rank): The average of 1/(position of first relevant result) across all queries, measuring how quickly users find their first relevant document.
- Precision@k: The proportion of the top-k retrieved documents that are actually relevant (e.g., P@10 of 0.5720 means 57.2% of top 10 results are relevant).
Results
We’ll look at 4 result sets
- first with a baseline embedding based search using a 768 dim vector embedding
- Using our LLM based search using a local model (qwen2.5:14b)
- running our LLM based search using Claude 4.5 as the LLM generating query sets.
- finally, allowing claude to optionally look for bi-grams (created a bi-gram index as well).
Baseline with embedding based search
I ran the benchmark using the sentence-transformers/all-mpnet-base-v2 embedding model for docs and queries (a 768-dimensional dense embedding model).
Note that it took a few mins to first generate embeddings for all documents in the dataset.
| Metric | mpnet-base-v2 baseline |
|---|---|
| NDCG@10 | 0.4725 |
| Recall@1000 | 0.3281 |
| MAP@1000 | 0.1331 |
| MRR@10 | 0.7244 |
LLM based search using Qwen 2.5 14b model
Qwen 2.5:14b running on-prem to see if the approach works with smaller, locally-hosted models:
| Metric | Qwen 2.5:14b | mpnet-base-v2 baseline | Difference |
|---|---|---|---|
| NDCG@10 | 0.407 | 0.4725 | -13.9% |
| Recall@1000 | 0.235 | 0.3281 | -28.3% |
| MAP@1000 | 0.076 | 0.1331 | -43.9% |
| MRR@10 | 0.626 | 0.7244 | -13.6% |
Hmm, the results are not great - also I’m running the qwen-2.5:14b model on my local machine (Macbook M4 Pro 24GB) and the latency for each query set generation is about 8s. Not great.
LLM based search using Claude 4.5
I had to run this benchmark 5 times because the results were reasonable and I didn’t believe it:
| Metric | Claude Sonnet 4.5 | mpnet-base-v2 baseline | Difference |
|---|---|---|---|
| NDCG@10 | 0.484 ± 0.019 | 0.4725 | +2.4% |
| Recall@1000 | 0.301 ± 0.004 | 0.3281 | -8.3% |
| MAP@1000 | 0.120 ± 0.003 | 0.1331 | -10.0% |
| MRR@10 | 0.714 ± 0.007 | 0.7244 | -1.4% |
NDCG@10 is 2.4% higher than the vector baseline, and MRR@10 is within 1.4%. For the top-ranked results (what users actually see), the inverted index approach performs remarkably close to the vector baseline. It was also surprising to see a relatively lower standard deviation which showed the approach was relatively stable between runs.
Also the results are much better than Qwen2.5:14b (not a fair comparison I know). Qwen’s also runs twice as slow despite being on-prem. Claude’s ability to generate creative, diverse keyword expansions directly impacts retrieval quality.
Claude 4.5 that can optionally generate bigrams
Bigrams are just 2 word sets so something like ‘machine learning’ or ‘Covid 19’. I added another index and allowed the LLM to generate bi-grams in its query sets. This resulted in query sets like the following
['nucleotide', 'polymorphism', 'mutation rate']
['dosage', 'outcomes', 'coronavirus disease']
These were the results with this approach (single run)
| Metric | Sonnet 4.5 (bigrams) | mpnet-base-v2 baseline | Diff |
|---|---|---|---|
| NDCG@10 | 0.5233 | 0.4725 | +0.0508 (+10.8%) |
| Recall@1000 | 0.2992 | 0.3281 | -0.0289 (-8.8%) |
| MAP@1000 | 0.1229 | 0.1331 | -0.0102 (-7.7%) |
| MRR@10 | 0.7910 | 0.7244 | +0.0666 (+9.2%) |
So recall didn’t improve much, instead it might’ve gone down a bit. NDCG and MRR did go up a little bit which makes sense.
Tradeoffs
Lower Recall
The gap between LLM based search and vector DBs is wide even at a 768 dim embedding. Even with our At Recall@1000, we’re 8.3% behind the vector baseline. So it looks like keyword matching, even with LLM-generated expansions, will miss semantic relationships that dense embeddings capture naturally.
Also, after running this experiment I checked Turbopuffer’s continuous recall numbers - they are able to get 90%+ for Recall@10 in production. That’s super impressive.
Latency
Latency is also higher. Across 5 runs with Claude Sonnet 4.5, processing 50 queries took an average of 244.9 seconds (0.20 QPS, ~4.9s per query). Most of this time is spent on LLM query generation - each query requires a Claude API call to generate the 10 keyword sets.
Breaking down the per-query latency:
- LLM query generation: ~3.5-4.0 seconds
- Inverted index search: ~0.01-0.02 seconds
- BM25 scoring: ~0.01-0.02 seconds
The search itself is fast. The LLM overhead is the bottleneck. With Qwen 2.5:14b, queries were even slower at ~8-9s each. There might be ways to speed up the local LLM execution but I just used ollama for now.
When This Makes Sense
This approach might work well when:
- You’re searching short, structured documents where keyword matching works well
- Quality matters more than raw speed for each individual query
- You want to avoid embedding infrastructure and its associated costs
- Your domain has precise terminology where keyword matching excels (medical, legal, technical docs)
It’s less suitable for:
- Long-form content where semantic understanding is critical
- High QPS workloads where sub-second latency is required
- Queries that require understanding context beyond keyword relationships
Conclusion
With 10 keyword sets generated from Claude 4.5 we managed to get performance comparable to the sentence-transformers/all-mpnet-base-v2 embedding model on our IR task. By varying configurations like more/fewer query sets, number of terms per set, etc this method might work well for many applications that prefer the simplicity of an inverted index, it might be a compelling alternative to vector databases.
All the code used in this experiment is available in my fork of the BEIR repo. The parts specific to the inverted index are available in the search_engine subdirectory.
